日本語での解説はNICT Press releaseに掲載。
The human brain comprises a number of white matter pathways involved with the communication between distant brain regions. Recent advances in diffusion magnetic resonance imaging (diffusion MRI) and tractography enable us to identify white matter pathways from the living human brain. This progress opens a new avenue for studying human white matter in relation to disease, development and behaviors.
Tractography estimates the trajectory of white matter pathways by tracking the orientation of diffusion MRI signal in each datapoint. Nowadays, a variety of tractography algorithms and softwares are widely used, each of which estimates white matter pathways based on different computational principles. In any algorithm, users are asked to select parameter values for performing the tractography. As a common practice, users chose a specific algorithm and a parameter set in order to analyze the diffusion MRI data. Choosing an algorithm and parameter settings poses a great challenge, as the results will significantly depend on that choice.
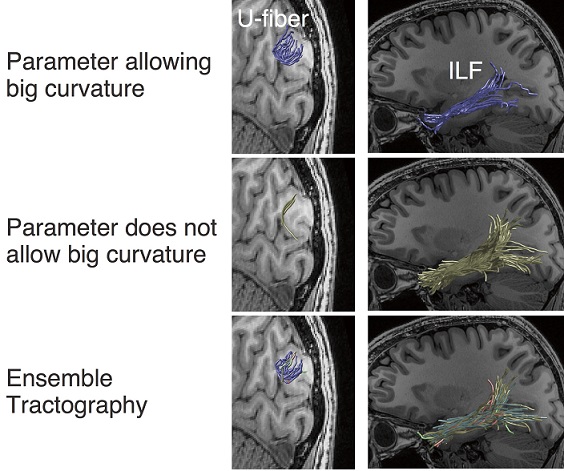
The top and middle panels of the figure below illustrates this problem. The panels show the estimates of white matter pathways using the same dataset with different parameters in tractography. In the top panel, a short-range pathway (U-fibers) is clearly identified whereas a long-range pathway (Inferior Longitudinal Fasciculus; ILF) is not well supported. In the middle panel, the ILF is clearly supported whereas the U-fiber is not. Thus, even when we analyze the same dataset, the estimates of the white matter pathways significantly depend on how we chose the parameters. Researchers have noted that this dependency makes it difficult for interpreting tractography results in a neurobiological context.
In this research, Hiromasa Takemura and collaborators (Franco Pestilli, Indiana Univ.; Cesar Caiafa, CONICET; Brian Wandell, Stanford Univ.) proposed a new method named “Ensemble Tractography”. In this method, white matter pathways are estimated using multiple algorithms and parameter settings, and the results are combined to create a super-set candidate model of white matter pathways. Next, Linear Fascicle Evaluation (LiFE; Pestilli et al., 2014, Nat Methods) is applied to select the valid pathways from the candidate model. Essentially, Ensemble Tractography enables us to take advantages from multiple methods; a super-set candidate is created, and LiFE selects the valid estimates from candidate.
The bottom panel in the figure describes the white matter pathways identified by Ensemble Tractography. Ensemble Tractography identified both U-fiber and the ILF, whereas conventional tractography with single parameter setting is biased to one or the other (top and middle panels). Moreover, Hiromasa and colleagues also showed that Ensemble Tractography provides higher model accuracy of white matter connections as compared with previous methods.
Ensemble Tractography addresses a widely recognized problem with tractography methods, namely, that the results depend significantly on the choice of algorithms and parameters. This advancement will improve the quality and reliability of tractography results, providing further opportunities for a more complete understanding about how information is transmitted in the human brain.
This paper appeared in the journal PLoS Computational Biology on February 4, 2016.
Full Reference:
“Ensemble Tractography”
Hiromasa Takemura, Cesar F. Caiafa, Brian A. Wandell, Franco Pestilli (2016) PLoS Computational Biology, 12(2): e1004692. doi:10.1371/journal.pcbi.1004692
URL: http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004692